about
launch
upload
download
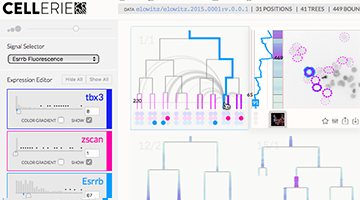

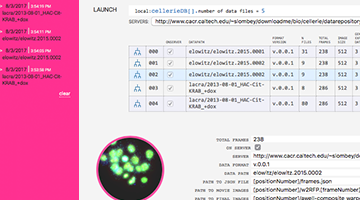
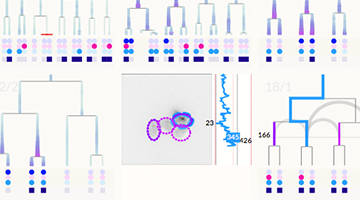
 QUICK LAUNCH: DEMO
QUICK LAUNCH: DEMO
{
"BIO_TREE_JSON_FORMAT": "v.0.0.1.2017.01.10",
"structureFields": [ "cellParent", "cellFrame", "bdryCells" ],
"nodeFields": [ "cellSig", "cellSize", "cellPos", "cellArea"],
"leafFields": [ "FISHcounts_Tbx3",
"FISHcounts_Esrrb",
"FISHcounts_Zscan4",
"FISH_prom_Act"
],
// ~~~~~~~ structure fields
"cellParent": [
-1, // node 0: root, no parent
0, // node 1: parent->0
0, // node 2: parent->0
1, // node 3: parent->1
1, // node 4: parent->1
2, // node 5: parent->2
2 // node 6: parent->2
],
"cellFrame": [
[1001, 1002, 1003, 1004], // node 0: born at 1001, splits after 1004
[1005, 1006, 1007, 1008, 1009], // node 1: born at 1005, splits after 1009
[1005, 1006, 1007, 1008], // node 2: born at 1005, splits after 1008
[1010, 1011, 1012, 1013], // node 3: born at 1010
[1010, 1011, 1012, 1013], // node 4: born at 1010
[1009, 1010, 1011, 1012, 1013], // node 5: born at 1009
[1009, 1010, 1011, 1012, 1013] // node 6: born at 1009
],
"bdryCells": [ 3, 4, 5, 6 ],
// ~~~~~~~ node fields
"cellSize": [
[[20,20], [21,20], [21,21], [22,22]], // node 0
[[19,20], [20,20], [21,21], [21,22], [21,22]], // node 1
[[18,18], [18,19], [19,19], [20,20]], // node 2
[[21,19], [21,20], [21,21], [22,21]], // node 3
[[20,21], [20,22], [20,22], [21,22]], // node 4
[[22,19], [22,20], [23,21], [24,21], [24,21]], // node 5
[[20,22], [21,22], [22,23], [23,23], [24,23]] // node 6
],
"cellPos": [
[[150,150], [151,150], [150,150], [151,151]], // node 0
[[140,151], [140,150], [139,150], [138,150], [137,150]], // node 1
[[160,150], [160,151], [161,152], [162,152]], // node 2
[[137,140], [137,139], [136,138], [135,137]], // node 3
[[137,160], [138,161], [139,162], [140,161]], // node 4
[[162,162], [162,163], [161,164], [162,165], [163,166]], // node 5
[[162,142], [163,142], [163,141], [162,140], [163,141]] // node 6
],
"cellArea": [
[311, 313, 314, 310], // node 0
[343, 318, 410, 342, 410], // node 1
[354, 403, 333, 413], // node 2
[297, 332, 414, 414], // node 3
[364, 345, 325, 395], // node 4
[378, 399, 413, 402, 383], // node 5
[313, 395, 362, 342, 361] // node 6
],
"cellSig": [
[103.2, 107.4, 103.2, 107.3], // node 0
[106.3, 104.8, 103.2, 109.3, 102.3], // node 1
[109.2, 109.3, 105.3, 103.8], // node 2
[102.1, 108.9, 108.1, 107.2], // node 3
[100.1, 103.2, 109.1, 108.3], // node 4
[100.8, 104.0, 103.3, 109.2, 106.3], // node 5
[108.3, 107.3, 100.0, 102.1, 104.3] // node 6
],
// ~~~~~~~ leaf node fields
"FISHcounts_Esrrb": [ 14, 21, 62, 47 ],
"FISHcounts_Tbx3": [ 0, 0, 1, 2 ],
"FISH_prom_Act": [ 0.51224, 2.1403, 0.81692, 0.15217 ],
"FISHcounts_Zscan4": [ 1, 0, 0, 11 ]
}
var myDataSet = {
formatversion: "v.0.0.1",
datapath: "data/my.experiment.0001",
files: [1,2,3,4,5],
pathToJsonFile: { path:"[positionNumber]/frames.json",
fixedWidth: false },
pathToMovieImages: { path:"[positionNumber]/frame.[frameNumber].jpg",
fixedWidth: false },
pathToFinalImages: { path:"[positionNumber]/finalframe.FISH.png",
fixedWidth: false },
imageSize: 512,
signalData: [ {arrayName:"cellSig", label:"Esrrb Fluorescence"} ],
totalFrames: 20,
geneExpressionData: [
{arrayName : "FISHcounts_Esrrb", label: "Esrrb"},
{arrayName : "FISHcounts_Zscan4", label: "zscan"},
{arrayName : "FISHcounts_Tbx3", label: "tbx3"}
]
};
cellerieDB.push(myDataSet);